
Coverage Report
File DbSourceProxyImpl.java
Coverage histogram
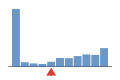
49% of files have more coverage
Code metrics
6
19
12
1
161
81
15
0.79
1.58
12
1.25
6
19
12
1
161
81
15
0.79
1.58
12
1.25
Classes
Class |
Line # |
Actions |
|||
|---|---|---|---|---|---|
| DbSourceProxyImpl | 36 | 19 | 15 | 0.3513513535.1% |
Class DbSourceProxyImpl
Class DbSourceProxyImpl |
19 |
15 |
0.3513513535.1% |
|
|---|---|---|---|---|
| DbSourceProxyImpl() DbSourceProxyImpl() | 4343 | 0.00 | 1.01 | -1.0 -1.0 - |
| getRawRecords() : StringBuffer getRawRecords() : StringBuffer | 5252 | 1.01 | 1.01 | 0.0 0.00% |
| queryInProgress() : boolean queryInProgress() : boolean | 6363 | 1.01 | 1.01 | 0.0 0.00% |
| startQuery() : void startQuery() : void | 7373 | 1.01 | 1.01 | 0.0 0.00% |
| stopQuery() : void stopQuery() : void | 8282 | 1.01 | 1.01 | 0.0 0.00% |
| parseResult(String) : AlignmentI parseResult(String) : AlignmentI | 9494 | 5.05 | 2.02 | 0.0 0.00% |
| getAccessionIdFromQuery(String) : String getAccessionIdFromQuery(String) : String | 111111 | 5.05 | 3.03 | 0.7777778 0.777777877.8% |
| getMaximumQueryCount() : int getMaximumQueryCount() : int | 126126 | 1.01 | 1.01 | 0.0 0.00% |
| isDnaCoding() : boolean isDnaCoding() : boolean | 135135 | 1.01 | 1.01 | 0.0 0.00% |
| isAlignmentSource() : boolean isAlignmentSource() : boolean | 144144 | 1.01 | 1.01 | 1.0 1.0100% |
| getDescription() : String getDescription() : String | 150150 | 1.01 | 1.01 | 0.0 0.00% |
| getFeatureColourScheme() : FeatureSettingsModelI getFeatureColourScheme() : FeatureSettingsModelI | 156156 | 1.01 | 1.01 | 1.0 1.0100% |
Contributing tests
This file is covered by 219 tests.
.
Contributing tests
|
0.24324325
|
jalview.ext.ensembl.EnsemblProteinTest.testGetAccesionIdFromQueryjalview.ext.ensembl.EnsemblProteinTest.testGetAccesionIdFromQuery | 1PASS | ||
|
0.13513513
|
jalview.io.CrossRef2xmlTests.openCrossrefsForEnsemblTwicejalview.io.CrossRef2xmlTests.openCrossrefsForEnsemblTwice | 1PASS | ||
|
0.08108108
|
jalview.project.Jalview2xmlTests.testCopyViewSettingsjalview.project.Jalview2xmlTests.testCopyViewSettings | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageOutputTestjalview.bin.CommandsTest.structureImageOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageOutputTestjalview.bin.CommandsTest.structureImageOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.08108108
|
jalview.ext.jmol.JmolViewerTest.testAddStrToSingleSeqViewJMoljalview.ext.jmol.JmolViewerTest.testAddStrToSingleSeqViewJMol | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest | 1PASS | ||
|
0.08108108
|
jalview.ws.dbsources.UniprotTest.testimportOfProblemEntriesjalview.ws.dbsources.UniprotTest.testimportOfProblemEntries | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest | 1PASS | ||
|
0.08108108
|
jalview.project.Jalview2xmlTests.viewRefPdbAnnotationjalview.project.Jalview2xmlTests.viewRefPdbAnnotation | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest | 1PASS | ||
|
0.08108108
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverExpandedviewsjalview.project.Jalview2xmlTests.testStoreAndRecoverExpandedviews | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest | 1PASS | ||
|
0.08108108
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverReferenceSeqSettingsjalview.project.Jalview2xmlTests.testStoreAndRecoverReferenceSeqSettings | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest | 1PASS | ||
|
0.08108108
|
jalview.ws.SequenceFetcherTest.testNoDuplicatesInFetchDbRefsjalview.ws.SequenceFetcherTest.testNoDuplicatesInFetchDbRefs | 1PASS | ||
|
0.08108108
|
jalview.gui.AnnotationLabelsTest2.testIdWidthChangesjalview.gui.AnnotationLabelsTest2.testIdWidthChanges | 1PASS | ||
|
0.08108108
|
jalview.project.Jalview2xmlTests.gatherViewsHerejalview.project.Jalview2xmlTests.gatherViewsHere | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageOutputTestjalview.bin.CommandsTest.structureImageOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest.structureImageOutputTestjalview.bin.CommandsTest.structureImageOutputTest | 1PASS | ||
|
0.08108108
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest | 1PASS | ||
|
0.054054055
|
jalview.datamodel.AlignmentTest.testAssertDatasetIsNormalisedjalview.datamodel.AlignmentTest.testAssertDatasetIsNormalised | 1PASS | ||
|
0.054054055
|
jalview.controller.AlignViewControllerTest.testSelectColumnsWithHighlightjalview.controller.AlignViewControllerTest.testSelectColumnsWithHighlight | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignViewportTest.testGetConsensusSeqjalview.gui.AlignViewportTest.testGetConsensusSeq | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTestjalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTest | 1PASS | ||
|
0.054054055
|
jalview.io.FeaturesFileTest.testParse_mixedJalviewGffjalview.io.FeaturesFileTest.testParse_mixedJalviewGff | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testMergeDatasetsforManyViewsjalview.project.Jalview2xmlTests.testMergeDatasetsforManyViews | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testAutoShowOverviewForLegacyProjectsjalview.project.Jalview2xmlTests.testAutoShowOverviewForLegacyProjects | 1PASS | ||
|
0.054054055
|
jalview.io.BackupFilesTest.backupsEnabledRollMaxTestjalview.io.BackupFilesTest.backupsEnabledRollMaxTest | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationColumnChooserTest.testResetjalview.gui.AnnotationColumnChooserTest.testReset | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignViewportTest.testDeregisterMapping_withReferencejalview.gui.AlignViewportTest.testDeregisterMapping_withReference | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTestjalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTest | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTestjalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTest | 1PASS | ||
|
0.054054055
|
jalview.gui.ScalePanelTest.testBuildPopupMenujalview.gui.ScalePanelTest.testBuildPopupMenu | 1PASS | ||
|
0.054054055
|
jalview.gui.FeatureSettingsTest.testSaveLoadjalview.gui.FeatureSettingsTest.testSaveLoad | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignFrameTest.testChangeColour_background_groupsAndThresholdsjalview.gui.AlignFrameTest.testChangeColour_background_groupsAndThresholds | 1PASS | ||
|
0.054054055
|
jalview.workers.AlignCalcManagerTest.testRemoveWorkerForAnnotationjalview.workers.AlignCalcManagerTest.testRemoveWorkerForAnnotation | 1PASS | ||
|
0.054054055
|
jalview.datamodel.AlignmentTest.testFindAnnotation_byCalcIdjalview.datamodel.AlignmentTest.testFindAnnotation_byCalcId | 1PASS | ||
|
0.054054055
|
jalview.datamodel.AlignmentTest.testDeleteHiddenSequencejalview.datamodel.AlignmentTest.testDeleteHiddenSequence | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testMergeDatasetsforViewsjalview.project.Jalview2xmlTests.testMergeDatasetsforViews | 1PASS | ||
|
0.054054055
|
jalview.io.SequenceAnnotationReportTest.testAppendFeature_colouredByAttributejalview.io.SequenceAnnotationReportTest.testAppendFeature_colouredByAttribute | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverGeneLocusjalview.project.Jalview2xmlTests.testStoreAndRecoverGeneLocus | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignViewportTest.testDeregisterMapping_onCloseViewjalview.gui.AlignViewportTest.testDeregisterMapping_onCloseView | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.054054055
|
jalview.util.CaseInsensitiveStringTest.testEqualsjalview.util.CaseInsensitiveStringTest.testEquals | 1PASS | ||
|
0.054054055
|
jalview.gui.ScalePanelTest.testSelectColumns_withHiddenjalview.gui.ScalePanelTest.testSelectColumns_withHidden | 1PASS | ||
|
0.054054055
|
jalview.datamodel.AlignmentTest.testCreateDatasetAlignmentWithMappedToSeqsjalview.datamodel.AlignmentTest.testCreateDatasetAlignmentWithMappedToSeqs | 1PASS | ||
|
0.054054055
|
jalview.io.TCoffeeScoreFileTest.testGetAsListjalview.io.TCoffeeScoreFileTest.testGetAsList | 1PASS | ||
|
0.054054055
|
jalview.schemes.ColourSchemesTest.testRegisterColourSchemejalview.schemes.ColourSchemesTest.testRegisterColourScheme | 1PASS | ||
|
0.054054055
|
jalview.io.vcf.VCFLoaderTest.testDoLoad_vepCsqjalview.io.vcf.VCFLoaderTest.testDoLoad_vepCsq | 1PASS | ||
|
0.054054055
|
jalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances_withSSUndefinedInBothSeqsjalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances_withSSUndefinedInBothSeqs | 1PASS | ||
|
0.054054055
|
jalview.io.AnnotatedPDBFileInputTest.testJalviewProjectRelocationAnnotationjalview.io.AnnotatedPDBFileInputTest.testJalviewProjectRelocationAnnotation | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignViewportTest.testSetSelectionGroupjalview.gui.AlignViewportTest.testSetSelectionGroup | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.054054055
|
jalview.gui.SeqCanvasTest.testCalculateWrappedGeometry_withAnnotationsjalview.gui.SeqCanvasTest.testCalculateWrappedGeometry_withAnnotations | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverColourThresholdsjalview.project.Jalview2xmlTests.testStoreAndRecoverColourThresholds | 1PASS | ||
|
0.054054055
|
jalview.io.BackupFilesTest.backupsEnabledSingleFileBackupTestjalview.io.BackupFilesTest.backupsEnabledSingleFileBackupTest | 1PASS | ||
|
0.054054055
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationChooserTest.testSelectType_showForSelectedjalview.gui.AnnotationChooserTest.testSelectType_showForSelected | 1PASS | ||
|
0.054054055
|
jalview.io.FeaturesFileTest.testPrintGffFormat_withFiltersjalview.io.FeaturesFileTest.testPrintGffFormat_withFilters | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignViewportTest.testSetGetHasSearchResultsjalview.gui.AlignViewportTest.testSetGetHasSearchResults | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationChooserTest.testDeselectType_showForSelectedjalview.gui.AnnotationChooserTest.testDeselectType_showForSelected | 1PASS | ||
|
0.054054055
|
jalview.structure.StructureSelectionManagerTest.testSetMapping_seqFeaturesjalview.structure.StructureSelectionManagerTest.testSetMapping_seqFeatures | 1PASS | ||
|
0.054054055
|
jalview.renderer.seqfeatures.FeatureRendererTest.testGetColourjalview.renderer.seqfeatures.FeatureRendererTest.testGetColour | 1PASS | ||
|
0.054054055
|
jalview.gui.QuitHandlerTest.testForceQuitjalview.gui.QuitHandlerTest.testForceQuit | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testStoreAndRestoreIDwidthAndAnnotationHeightjalview.project.Jalview2xmlTests.testStoreAndRestoreIDwidthAndAnnotationHeight | 1PASS | ||
|
0.054054055
|
jalview.io.SequenceAnnotationReportTest.testAppendFeature_statusjalview.io.SequenceAnnotationReportTest.testAppendFeature_status | 1PASS | ||
|
0.054054055
|
jalview.datamodel.AlignmentTest.testDeleteAllAnnotations_excludingAutocalculatedjalview.datamodel.AlignmentTest.testDeleteAllAnnotations_excludingAutocalculated | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationLabelsTest2.testIdWidthChangesjalview.gui.AnnotationLabelsTest2.testIdWidthChanges | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTestjalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTest | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationLabelsTest2.testIdWidthNoChangesjalview.gui.AnnotationLabelsTest2.testIdWidthNoChanges | 1PASS | ||
|
0.054054055
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.054054055
|
jalview.datamodel.AlignmentTest.testCreateDataset_updateDbrefMappingsjalview.datamodel.AlignmentTest.testCreateDataset_updateDbrefMappings | 1PASS | ||
|
0.054054055
|
jalview.io.BackupFilesTest.backupsEnabledNoRollMaxTestjalview.io.BackupFilesTest.backupsEnabledNoRollMaxTest | 1PASS | ||
|
0.054054055
|
jalview.gui.StructureChooserTest.displayTDBQueryTestjalview.gui.StructureChooserTest.displayTDBQueryTest | 1PASS | ||
|
0.054054055
|
jalview.io.SequenceAnnotationReportTest.testAppendFeature_noScorejalview.io.SequenceAnnotationReportTest.testAppendFeature_noScore | 1PASS | ||
|
0.054054055
|
jalview.io.ScoreMatrixFileTest.testParseMatrix_aaindex_tooFewColumnsjalview.io.ScoreMatrixFileTest.testParseMatrix_aaindex_tooFewColumns | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.054054055
|
jalview.io.gff.ExonerateHelperTest.testAddExonerateGffToAlignmentjalview.io.gff.ExonerateHelperTest.testAddExonerateGffToAlignment | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationChooserTest.testDeselectType_hideForSelectedjalview.gui.AnnotationChooserTest.testDeselectType_hideForSelected | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignFrameTest.testNewView_dsRefPreservedjalview.gui.AlignFrameTest.testNewView_dsRefPreserved | 1PASS | ||
|
0.054054055
|
jalview.datamodel.AlignmentTest.testEqualsjalview.datamodel.AlignmentTest.testEquals | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.noDuplicatePdbMappingsMadejalview.project.Jalview2xmlTests.noDuplicatePdbMappingsMade | 1PASS | ||
|
0.054054055
|
jalview.schemes.ColourSchemesTest.testGetColourSchemejalview.schemes.ColourSchemesTest.testGetColourScheme | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignViewportTest.testGetSelectionAsNewSequences_withContactMatricesjalview.gui.AlignViewportTest.testGetSelectionAsNewSequences_withContactMatrices | 1PASS | ||
|
0.054054055
|
jalview.io.SequenceAnnotationReportTest.testAppendFeature_disulfideBondjalview.io.SequenceAnnotationReportTest.testAppendFeature_disulfideBond | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignFrameTest.testNewView_colourThresholdsjalview.gui.AlignFrameTest.testNewView_colourThresholds | 1PASS | ||
|
0.054054055
|
jalview.datamodel.AlignmentTest.testDeleteSequenceBySeqjalview.datamodel.AlignmentTest.testDeleteSequenceBySeq | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testPAEsaveRestorejalview.project.Jalview2xmlTests.testPAEsaveRestore | 1PASS | ||
|
0.054054055
|
jalview.gui.QuitHandlerTest.testSavedAlignmentChangesjalview.gui.QuitHandlerTest.testSavedAlignmentChanges | 1PASS | ||
|
0.054054055
|
jalview.gui.PairwiseAlignmentPanelTest.testConstructor_noSelectionGroupjalview.gui.PairwiseAlignmentPanelTest.testConstructor_noSelectionGroup | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationChooserTest.testSelectType_showForAlljalview.gui.AnnotationChooserTest.testSelectType_showForAll | 1PASS | ||
|
0.054054055
|
jalview.io.FeaturesFileTest.testParsejalview.io.FeaturesFileTest.testParse | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationChooserTest.testBuildShowHidePaneljalview.gui.AnnotationChooserTest.testBuildShowHidePanel | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testPcaViewAssociationjalview.project.Jalview2xmlTests.testPcaViewAssociation | 1PASS | ||
|
0.054054055
|
jalview.renderer.ScaleRendererTest.testCalculateMarksjalview.renderer.ScaleRendererTest.testCalculateMarks | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverGroupRepSeqsjalview.project.Jalview2xmlTests.testStoreAndRecoverGroupRepSeqs | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverNoOverviewjalview.project.Jalview2xmlTests.testStoreAndRecoverNoOverview | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testColourByAnnotScoresjalview.project.Jalview2xmlTests.testColourByAnnotScores | 1PASS | ||
|
0.054054055
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModeljalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModel | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationLabelsTest2.testIdWidthNoChangesjalview.gui.AnnotationLabelsTest2.testIdWidthNoChanges | 1PASS | ||
|
0.054054055
|
jalview.gui.QuitHandlerTest.testNoGUIUnsavedChangesjalview.gui.QuitHandlerTest.testNoGUIUnsavedChanges | 1PASS | ||
|
0.054054055
|
jalview.io.TCoffeeScoreFileTest.testHeightAndWidthWithResidueNumbersjalview.io.TCoffeeScoreFileTest.testHeightAndWidthWithResidueNumbers | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.054054055
|
jalview.gui.QuitHandlerTest.testSavedProjectChangesjalview.gui.QuitHandlerTest.testSavedProjectChanges | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.054054055
|
jalview.schemes.ClustalxColourSchemeTest.testFindColour_ignoreGapsjalview.schemes.ClustalxColourSchemeTest.testFindColour_ignoreGaps | 1PASS | ||
|
0.054054055
|
jalview.schemes.PIDColourSchemeTest.testFindColour_ignoreGapsjalview.schemes.PIDColourSchemeTest.testFindColour_ignoreGaps | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.054054055
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.054054055
|
jalview.gui.SeqPanelTest.testSetStatusReturnsNearestResiduePositionjalview.gui.SeqPanelTest.testSetStatusReturnsNearestResiduePosition | 1PASS | ||
|
0.054054055
|
jalview.gui.QuitHandlerTest.testUnsavedChangesjalview.gui.QuitHandlerTest.testUnsavedChanges | 1PASS | ||
|
0.054054055
|
jalview.gui.ProgressBarTest.testSetProgressBarjalview.gui.ProgressBarTest.testSetProgressBar | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignmentPanelTest.testCalculateIdWidth_noArgsjalview.gui.AlignmentPanelTest.testCalculateIdWidth_noArgs | 1PASS | ||
|
0.054054055
|
jalview.renderer.seqfeatures.FeatureRendererTest.testFindComplementFeaturesAtResiduejalview.renderer.seqfeatures.FeatureRendererTest.testFindComplementFeaturesAtResidue | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverAnnotationRowElementColoursjalview.project.Jalview2xmlTests.testStoreAndRecoverAnnotationRowElementColours | 1PASS | ||
|
0.054054055
|
jalview.io.FeaturesFileTest.testPrintGffFormatjalview.io.FeaturesFileTest.testPrintGffFormat | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationChooserTest.testDeselectType_hideForAlljalview.gui.AnnotationChooserTest.testDeselectType_hideForAll | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignmentPanelTest.testSetOverviewTitle_automaticOverviewjalview.gui.AlignmentPanelTest.testSetOverviewTitle_automaticOverview | 1PASS | ||
|
0.054054055
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModel_HiddenColumnsjalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModel_HiddenColumns | 1PASS | ||
|
0.054054055
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.054054055
|
jalview.ext.jmol.JmolParserTest.testAlignmentLoaderjalview.ext.jmol.JmolParserTest.testAlignmentLoader | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignmentPanelTest.testSetScrollValuesjalview.gui.AlignmentPanelTest.testSetScrollValues | 1PASS | ||
|
0.054054055
|
jalview.io.SequenceAnnotationReportTest.testAppendFeature_stripHtmljalview.io.SequenceAnnotationReportTest.testAppendFeature_stripHtml | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignViewportTest.testUpdateConservation_qualityOnlyjalview.gui.AlignViewportTest.testUpdateConservation_qualityOnly | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testTCoffeeScoresjalview.project.Jalview2xmlTests.testTCoffeeScores | 1PASS | ||
|
0.054054055
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.054054055
|
jalview.gui.PopupMenuTest.testBuildLinkMenujalview.gui.PopupMenuTest.testBuildLinkMenu | 1PASS | ||
|
0.054054055
|
jalview.datamodel.AlignmentTest.testDeleteSequenceByIndexjalview.datamodel.AlignmentTest.testDeleteSequenceByIndex | 1PASS | ||
|
0.054054055
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFindFeatureAt_PointFeaturejalview.analysis.scoremodels.FeatureDistanceModelTest.testFindFeatureAt_PointFeature | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignViewportTest.testShowOrDontShowOccupancyjalview.gui.AlignViewportTest.testShowOrDontShowOccupancy | 1PASS | ||
|
0.054054055
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFindDistances_withParamsjalview.analysis.scoremodels.FeatureDistanceModelTest.testFindDistances_withParams | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest | 1PASS | ||
|
0.054054055
|
jalview.gui.AnnotationChooserTest.testSelectType_hideForSelectedjalview.gui.AnnotationChooserTest.testSelectType_hideForSelected | 1PASS | ||
|
0.054054055
|
jalview.ext.jmol.JmolParserTest.testFileParserjalview.ext.jmol.JmolParserTest.testFileParser | 1PASS | ||
|
0.054054055
|
jalview.gui.DesktopTests.testInternalCopyPastejalview.gui.DesktopTests.testInternalCopyPaste | 1PASS | ||
|
0.054054055
|
jalview.io.ScoreMatrixFileTest.testParseMatrix_ncbiBadFloatjalview.io.ScoreMatrixFileTest.testParseMatrix_ncbiBadFloat | 1PASS | ||
|
0.054054055
|
jalview.ext.jmol.JmolViewerTest.testSingleSeqViewJMoljalview.ext.jmol.JmolViewerTest.testSingleSeqViewJMol | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testRNAStructureRecoveryjalview.project.Jalview2xmlTests.testRNAStructureRecovery | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTestjalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTest | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testSaveLoadFeatureColoursAndFiltersjalview.project.Jalview2xmlTests.testSaveLoadFeatureColoursAndFilters | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.054054055
|
jalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances_withSSUndefinedInEitherOneSeqjalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances_withSSUndefinedInEitherOneSeq | 1PASS | ||
|
0.054054055
|
jalview.gui.SeqPanelTest.testFindMousePosition_wrapped_scales_longSequencejalview.gui.SeqPanelTest.testFindMousePosition_wrapped_scales_longSequence | 1PASS | ||
|
0.054054055
|
jalview.io.TCoffeeScoreFileTest.testGetAsArrayjalview.io.TCoffeeScoreFileTest.testGetAsArray | 1PASS | ||
|
0.054054055
|
jalview.ext.pymol.PymolManagerTest.testGetPostRequestjalview.ext.pymol.PymolManagerTest.testGetPostRequest | 1PASS | ||
|
0.054054055
|
jalview.gui.StructureChooserTest.populateFilterComboBoxTestjalview.gui.StructureChooserTest.populateFilterComboBoxTest | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.054054055
|
jalview.io.AnnotationExporterTest.testAnnotationExportAsCSVjalview.io.AnnotationExporterTest.testAnnotationExportAsCSV | 1PASS | ||
|
0.054054055
|
jalview.gui.AlignViewportTest.testSetGlobalColourSchemejalview.gui.AlignViewportTest.testSetGlobalColourScheme | 1PASS | ||
|
0.054054055
|
jalview.io.FeaturesFileTest.testParse_pureGff3jalview.io.FeaturesFileTest.testParse_pureGff3 | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverPDBEntryjalview.project.Jalview2xmlTests.testStoreAndRecoverPDBEntry | 1PASS | ||
|
0.054054055
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.054054055
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverOverviewjalview.project.Jalview2xmlTests.testStoreAndRecoverOverview | 1PASS | ||
|
0.054054055
|
jalview.gui.QuitHandlerTest.testWaitForSaveQuitjalview.gui.QuitHandlerTest.testWaitForSaveQuit | 1PASS | ||
|
0.054054055
|
jalview.math.SparseMatrixTest.testConstructorjalview.math.SparseMatrixTest.testConstructor | 1PASS | ||
|
0.054054055
|
jalview.datamodel.AlignmentTest.testCreateDatasetAlignmentjalview.datamodel.AlignmentTest.testCreateDatasetAlignment | 1PASS | ||
|
0.054054055
|
jalview.renderer.seqfeatures.FeatureRendererTest.testFindAllFeaturesjalview.renderer.seqfeatures.FeatureRendererTest.testFindAllFeatures | 1PASS | ||
|
0.054054055
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.054054055
|
jalview.io.SequenceAnnotationReportTest.testAppendFeature_virtualFeaturejalview.io.SequenceAnnotationReportTest.testAppendFeature_virtualFeature | 1PASS | ||
|
0.054054055
|
jalview.analysis.AverageDistanceEngineTest.testUPGMAEnginejalview.analysis.AverageDistanceEngineTest.testUPGMAEngine | 1PASS | ||
|
0.054054055
|
jalview.io.JSONFileTest.testBioJSONRoundTripWithColourSchemeNonejalview.io.JSONFileTest.testBioJSONRoundTripWithColourSchemeNone | 1PASS | ||
|
0.054054055
|
jalview.io.BackupFilesTest.backupsEnabledReverseRollMaxTestjalview.io.BackupFilesTest.backupsEnabledReverseRollMaxTest | 1PASS | ||
|
0.054054055
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModel_hiddenFirstColumnjalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModel_hiddenFirstColumn | 1PASS | ||
|
0.054054055
|
jalview.gui.ColourMenuHelperTest.testAddMenuItems_nucleotidejalview.gui.ColourMenuHelperTest.testAddMenuItems_nucleotide | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblGeneTest.testGetFeatureColourSchemejalview.ext.ensembl.EnsemblGeneTest.testGetFeatureColourScheme | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblProteinTest.testIsValidReferencejalview.ext.ensembl.EnsemblProteinTest.testIsValidReference | 1PASS | ||
|
0.027027028
|
jalview.ws.dbsources.RfamSeedTest.testGetURLjalview.ws.dbsources.RfamSeedTest.testGetURL | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblCdsTest.testGetIdentifyingFeaturesjalview.ext.ensembl.EnsemblCdsTest.testGetIdentifyingFeatures | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblGeneTest.testGetGenomicRangesFromFeatures_ncRNA_gene_reverseStrandjalview.ext.ensembl.EnsemblGeneTest.testGetGenomicRangesFromFeatures_ncRNA_gene_reverseStrand | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblCdsTest.testRetainFeaturejalview.ext.ensembl.EnsemblCdsTest.testRetainFeature | 1PASS | ||
|
0.027027028
|
jalview.ws.dbsources.RfamFullTest.testGetURLjalview.ws.dbsources.RfamFullTest.testGetURL | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblXrefTest.testGetCrossReferencesjalview.ext.ensembl.EnsemblXrefTest.testGetCrossReferences | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblGenomeTest.testGetGenomicRangesFromFeaturesjalview.ext.ensembl.EnsemblGenomeTest.testGetGenomicRangesFromFeatures | 1PASS | ||
|
0.027027028
|
jalview.ws.dbsources.PfamFullTest.testGetURLjalview.ws.dbsources.PfamFullTest.testGetURL | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblGenomeTest.testRetainFeaturejalview.ext.ensembl.EnsemblGenomeTest.testRetainFeature | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblCdnaTest.testRetainFeaturejalview.ext.ensembl.EnsemblCdnaTest.testRetainFeature | 1PASS | ||
|
0.027027028
|
jalview.ws.dbsources.UniprotTest.testGetUniprotEntryDescriptionjalview.ws.dbsources.UniprotTest.testGetUniprotEntryDescription | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblCdnaTest.testGetGenomicRangesFromFeaturesjalview.ext.ensembl.EnsemblCdnaTest.testGetGenomicRangesFromFeatures | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblGeneTest.testRetainFeaturejalview.ext.ensembl.EnsemblGeneTest.testRetainFeature | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblCdnaTest.testIsValidReferencejalview.ext.ensembl.EnsemblCdnaTest.testIsValidReference | 1PASS | ||
|
0.027027028
|
jalview.ws.dbsources.UniprotTest.testGetUniprotEntriesjalview.ws.dbsources.UniprotTest.testGetUniprotEntries | 1PASS | ||
|
0.027027028
|
jalview.ws.seqfetcher.DbRefFetcherTest.testStandardProtDbsjalview.ws.seqfetcher.DbRefFetcherTest.testStandardProtDbs | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblGeneTest.testGetTranscriptFeaturesjalview.ext.ensembl.EnsemblGeneTest.testGetTranscriptFeatures | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblGeneTest.testGetGenomicRangesFromFeaturesjalview.ext.ensembl.EnsemblGeneTest.testGetGenomicRangesFromFeatures | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblCdsTest.testIsValidReferencejalview.ext.ensembl.EnsemblCdsTest.testIsValidReference | 1PASS | ||
|
0.027027028
|
jalview.ws.dbsources.UniprotTest.testGetUniprotSequencejalview.ws.dbsources.UniprotTest.testGetUniprotSequence | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblCdnaTest.testGetGenomicRangesFromFeatures_mixedStrandjalview.ext.ensembl.EnsemblCdnaTest.testGetGenomicRangesFromFeatures_mixedStrand | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblGenomeTest.testGetIdentifyingFeaturesjalview.ext.ensembl.EnsemblGenomeTest.testGetIdentifyingFeatures | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblCdnaTest.testGetGenomicRangesFromFeatures_reverseStrandjalview.ext.ensembl.EnsemblCdnaTest.testGetGenomicRangesFromFeatures_reverseStrand | 1PASS | ||
|
0.027027028
|
jalview.ws.dbsources.PfamSeedTest.testGetURLjalview.ws.dbsources.PfamSeedTest.testGetURL | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblGeneTest.testGetIdentifyingFeaturesjalview.ext.ensembl.EnsemblGeneTest.testGetIdentifyingFeatures | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblCdnaTest.testGetIdentifyingFeaturesjalview.ext.ensembl.EnsemblCdnaTest.testGetIdentifyingFeatures | 1PASS | ||
|
0.027027028
|
jalview.ws.dbsources.UniprotTest.testGetUniprotEntryIdjalview.ws.dbsources.UniprotTest.testGetUniprotEntryId | 1PASS | ||
|
0.027027028
|
jalview.ext.ensembl.EnsemblCdsTest.testGetGenomicRangesFromFeaturesjalview.ext.ensembl.EnsemblCdsTest.testGetGenomicRangesFromFeatures | 1PASS |
Source view
| 1 | /* | ||||||
| 2 | * Jalview - A Sequence Alignment Editor and Viewer ($$Version-Rel$$) | ||||||
| 3 | * Copyright (C) $$Year-Rel$$ The Jalview Authors | ||||||
| 4 | * | ||||||
| 5 | * This file is part of Jalview. | ||||||
| 6 | * | ||||||
| 7 | * Jalview is free software: you can redistribute it and/or | ||||||
| 8 | * modify it under the terms of the GNU General Public License | ||||||
| 9 | * as published by the Free Software Foundation, either version 3 | ||||||
| 10 | * of the License, or (at your option) any later version. | ||||||
| 11 | * | ||||||
| 12 | * Jalview is distributed in the hope that it will be useful, but | ||||||
| 13 | * WITHOUT ANY WARRANTY; without even the implied warranty | ||||||
| 14 | * of MERCHANTABILITY or FITNESS FOR A PARTICULAR | ||||||
| 15 | * PURPOSE. See the GNU General Public License for more details. | ||||||
| 16 | * | ||||||
| 17 | * You should have received a copy of the GNU General Public License | ||||||
| 18 | * along with Jalview. If not, see <http://www.gnu.org/licenses/>. | ||||||
| 19 | * The Jalview Authors are detailed in the 'AUTHORS' file. | ||||||
| 20 | */ | ||||||
| 21 | package jalview.ws.seqfetcher; | ||||||
| 22 | |||||||
| 23 | import jalview.api.FeatureSettingsModelI; | ||||||
| 24 | import jalview.datamodel.AlignmentI; | ||||||
| 25 | import jalview.io.DataSourceType; | ||||||
| 26 | import jalview.io.FileFormatI; | ||||||
| 27 | import jalview.io.FormatAdapter; | ||||||
| 28 | import jalview.io.IdentifyFile; | ||||||
| 29 | |||||||
| 30 | /** | ||||||
| 31 | * common methods for implementations of the DbSourceProxy interface. | ||||||
| 32 | * | ||||||
| 33 | * @author JimP | ||||||
| 34 | * | ||||||
| 35 | */ | ||||||
|
|||||||
| 36 | public abstract class DbSourceProxyImpl implements DbSourceProxy | ||||||
| 37 | { | ||||||
| 38 | |||||||
| 39 | boolean queryInProgress = false; | ||||||
| 40 | |||||||
| 41 | protected StringBuffer results = null; | ||||||
| 42 | |||||||
|
|||||||
| 43 | 219 |  public DbSourceProxyImpl()... public DbSourceProxyImpl()... |
|||||
| 44 | { | ||||||
| 45 | } | ||||||
| 46 | |||||||
| 47 | /* | ||||||
| 48 | * (non-Javadoc) | ||||||
| 49 | * | ||||||
| 50 | * @see jalview.ws.DbSourceProxy#getRawRecords() | ||||||
| 51 | */ | ||||||
|
|||||||
| 52 | 0 |  @Override... @Override... |
|||||
| 53 | public StringBuffer getRawRecords() | ||||||
| 54 | { | ||||||
| 55 | 0 | return results; | |||||
| 56 | } | ||||||
| 57 | |||||||
| 58 | /* | ||||||
| 59 | * (non-Javadoc) | ||||||
| 60 | * | ||||||
| 61 | * @see jalview.ws.DbSourceProxy#queryInProgress() | ||||||
| 62 | */ | ||||||
|
|||||||
| 63 | 0 |  @Override... @Override... |
|||||
| 64 | public boolean queryInProgress() | ||||||
| 65 | { | ||||||
| 66 | 0 | return queryInProgress; | |||||
| 67 | } | ||||||
| 68 | |||||||
| 69 | /** | ||||||
| 70 | * call to set the queryInProgress flag | ||||||
| 71 | * | ||||||
| 72 | */ | ||||||
|
|||||||
| 73 | 0 |  protected void startQuery()... protected void startQuery()... |
|||||
| 74 | { | ||||||
| 75 | 0 | queryInProgress = true; | |||||
| 76 | } | ||||||
| 77 | |||||||
| 78 | /** | ||||||
| 79 | * call to clear the queryInProgress flag | ||||||
| 80 | * | ||||||
| 81 | */ | ||||||
|
|||||||
| 82 | 0 |  protected void stopQuery()... protected void stopQuery()... |
|||||
| 83 | { | ||||||
| 84 | 0 | queryInProgress = false; | |||||
| 85 | } | ||||||
| 86 | |||||||
| 87 | /** | ||||||
| 88 | * create an alignment from raw text file... | ||||||
| 89 | * | ||||||
| 90 | * @param result | ||||||
| 91 | * @return null or a valid alignment | ||||||
| 92 | * @throws Exception | ||||||
| 93 | */ | ||||||
|
|||||||
| 94 | 0 |  protected AlignmentI parseResult(String result) throws Exception... protected AlignmentI parseResult(String result) throws Exception... |
|||||
| 95 | { | ||||||
| 96 | 0 | AlignmentI sequences = null; | |||||
| 97 | 0 | FileFormatI format = new IdentifyFile().identify(result, | |||||
| 98 | DataSourceType.PASTE); | ||||||
| 99 | 0 | if (format != null) | |||||
| 100 | { | ||||||
| 101 | 0 | sequences = new FormatAdapter().readFile(result.toString(), | |||||
| 102 | DataSourceType.PASTE, format); | ||||||
| 103 | } | ||||||
| 104 | 0 | return sequences; | |||||
| 105 | } | ||||||
| 106 | |||||||
| 107 | /** | ||||||
| 108 | * Returns the first accession id in the query (up to the first accession id | ||||||
| 109 | * separator), or the whole query if there is no separator or it is not found | ||||||
| 110 | */ | ||||||
|
|||||||
| 111 | 6 |  @Override... @Override... |
|||||
| 112 | public String getAccessionIdFromQuery(String query) | ||||||
| 113 | { | ||||||
| 114 | 6 | String sep = getAccessionSeparator(); | |||||
| 115 | 6 | if (sep == null) | |||||
| 116 | { | ||||||
| 117 | 0 | return query; | |||||
| 118 | } | ||||||
| 119 | 6 | int sepPos = query.indexOf(sep); | |||||
| 120 | 6 | return sepPos == -1 ? query : query.substring(0, sepPos); | |||||
| 121 | } | ||||||
| 122 | |||||||
| 123 | /** | ||||||
| 124 | * Default is only one accession id per query - override if more are allowed. | ||||||
| 125 | */ | ||||||
|
|||||||
| 126 | 0 |  @Override... @Override... |
|||||
| 127 | public int getMaximumQueryCount() | ||||||
| 128 | { | ||||||
| 129 | 0 | return 1; | |||||
| 130 | } | ||||||
| 131 | |||||||
| 132 | /** | ||||||
| 133 | * Returns false - override to return true for DNA coding data sources | ||||||
| 134 | */ | ||||||
|
|||||||
| 135 | 0 |  @Override... @Override... |
|||||
| 136 | public boolean isDnaCoding() | ||||||
| 137 | { | ||||||
| 138 | 0 | return false; | |||||
| 139 | } | ||||||
| 140 | |||||||
| 141 | /** | ||||||
| 142 | * Answers false - override as required in subclasses | ||||||
| 143 | */ | ||||||
|
|||||||
| 144 | 13475 |  @Override... @Override... |
|||||
| 145 | public boolean isAlignmentSource() | ||||||
| 146 | { | ||||||
| 147 | 13475 | return false; | |||||
| 148 | } | ||||||
| 149 | |||||||
|
|||||||
| 150 | 0 |  @Override... @Override... |
|||||
| 151 | public String getDescription() | ||||||
| 152 | { | ||||||
| 153 | 0 | return ""; | |||||
| 154 | } | ||||||
| 155 | |||||||
|
|||||||
| 156 | 2 |  @Override... @Override... |
|||||
| 157 | public FeatureSettingsModelI getFeatureColourScheme() | ||||||
| 158 | { | ||||||
| 159 | 2 | return null; | |||||
| 160 | } | ||||||
| 161 | } |