
Coverage Report
File SequenceCursor.java
Coverage histogram
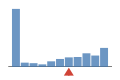
37% of files have more coverage
Code metrics
6
19
5
1
145
55
8
0.42
3.8
5
1.6
6
19
5
1
145
55
8
0.42
3.8
5
1.6
Classes
| Class | Line # | Actions | |||
|---|---|---|---|---|---|
| SequenceCursor | 27 | 19 | 8 | 0.660% |
Class SequenceCursor
|
Class SequenceCursor |
Line # 27 |
19 |
8 |
0.660% |
|---|---|---|---|---|
| SequenceCursor(SequenceI,int,int,int) SequenceCursor(SequenceI,int,int,int) | 7373 | 1.01 | 1.01 | 1.0 1.0100% |
| SequenceCursor(SequenceI,int,int,int,int,int) SequenceCursor(SequenceI,int,int,int,int,int) | 9797 | 6.06 | 1.01 | 1.0 1.0100% |
| hashCode() : int hashCode() : int | 108108 | 6.06 | 2.02 | 0.0 0.00% |
| equals(Object) : boolean equals(Object) : boolean | 125125 | 4.04 | 2.02 | 0.6666667 0.666666766.7% |
| toString() : String toString() : String | 137137 | 2.02 | 2.02 | 0.75 0.7575% |
Contributing tests
This file is covered by 300 tests.
.
Contributing tests
|
0.6
|
jalview.datamodel.SequenceTest.testFindPositionjalview.datamodel.SequenceTest.testFindPosition | 1PASS | ||
|
0.46666667
|
jalview.datamodel.SequenceTest.testFindPosition_withCursorAndEditsjalview.datamodel.SequenceTest.testFindPosition_withCursorAndEdits | 1PASS | ||
|
0.43333334
|
jalview.datamodel.SequenceTest.testFindPosition_withCursorjalview.datamodel.SequenceTest.testFindPosition_withCursor | 1PASS | ||
|
0.3
|
jalview.datamodel.SequenceTest.testFindIndex_withCursorjalview.datamodel.SequenceTest.testFindIndex_withCursor | 1PASS | ||
|
0.23333333
|
jalview.commands.TrimRegionCommandTest.testTrimLeft_withUndoAndRedojalview.commands.TrimRegionCommandTest.testTrimLeft_withUndoAndRedo | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequenceAndRequireAligned_ShortSeqRemovedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequenceAndRequireAligned_ShortSeqRemoved | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverHmmProfilejalview.project.Jalview2xmlTests.testStoreAndRecoverHmmProfile | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances_withSSUndefinedInEitherOneSeqjalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances_withSSUndefinedInEitherOneSeq | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverReferenceSeqSettingsjalview.project.Jalview2xmlTests.testStoreAndRecoverReferenceSeqSettings | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testReplace_withGapsjalview.commands.EditCommandTest.testReplace_withGaps | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_multipleFeaturesAtPositionNoTransparencyjalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_multipleFeaturesAtPositionNoTransparency | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_RegionIncludingGaps_RegionSubmittedWithoutGapsjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_RegionIncludingGaps_RegionSubmittedWithoutGaps | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_contactFeaturejalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_contactFeature | 1PASS | ||
|
0.23333333
|
mc_view.PDBfileTest.testParse_withJmolAddAlignmentAnnotationsmc_view.PDBfileTest.testParse_withJmolAddAlignmentAnnotations | 1PASS | ||
|
0.23333333
|
jalview.ext.jmol.JmolParserTest.testAlignmentLoaderjalview.ext.jmol.JmolParserTest.testAlignmentLoader | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.gui.AlignmentPanelTest.testPrintWrappedAlignmentjalview.gui.AlignmentPanelTest.testPrintWrappedAlignment | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testCutjalview.commands.EditCommandTest.testCut | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFindDistances_withParamsjalview.analysis.scoremodels.FeatureDistanceModelTest.testFindDistances_withParams | 1PASS | ||
|
0.23333333
|
jalview.io.StockholmFileTest.curlyWUSSsecondaryStructureForRNASequencejalview.io.StockholmFileTest.curlyWUSSsecondaryStructureForRNASequence | 1PASS | ||
|
0.23333333
|
jalview.analysis.FinderTest.testFind_findAllInSelectionWithShortSequencejalview.analysis.FinderTest.testFind_findAllInSelectionWithShortSequence | 1PASS | ||
|
0.23333333
|
jalview.gui.AnnotationLabelsTest.testDrawLabelsjalview.gui.AnnotationLabelsTest.testDrawLabels | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentSorterTest.testSortByFeature_scorejalview.analysis.AlignmentSorterTest.testSortByFeature_score | 1PASS | ||
|
0.23333333
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureRendererTest.testFindComplementFeaturesAtResiduejalview.renderer.seqfeatures.FeatureRendererTest.testFindComplementFeaturesAtResidue | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.ext.jmol.JmolViewerTest.testSingleSeqViewJMoljalview.ext.jmol.JmolViewerTest.testSingleSeqViewJMol | 1PASS | ||
|
0.23333333
|
jalview.io.AnnotationFileIOTest.exampleAnnotationFileIOjalview.io.AnnotationFileIOTest.exampleAnnotationFileIO | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_network_structureOpeningArgsTestjalview.bin.CommandsTest2.do_network_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceTest.testFindIndexjalview.datamodel.SequenceTest.testFindIndex | 1PASS | ||
|
0.23333333
|
jalview.io.AnnotationFileIOTest.testCalcIdRestorejalview.io.AnnotationFileIOTest.testCalcIdRestore | 1PASS | ||
|
0.23333333
|
jalview.io.AnnotatedPDBFileInputTest.testJalviewProjectRelocationAnnotationjalview.io.AnnotatedPDBFileInputTest.testJalviewProjectRelocationAnnotation | 1PASS | ||
|
0.23333333
|
jalview.io.AnnotationFileIOTest.testProviderRestorejalview.io.AnnotationFileIOTest.testProviderRestore | 1PASS | ||
|
0.23333333
|
jalview.analysis.FinderTest.testFind_regexjalview.analysis.FinderTest.testFind_regex | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.retrieveResult_verifySequencesAlignedjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.retrieveResult_verifySequencesAligned | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_SequenceTooShort_InputInvalidExceptionjalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_SequenceTooShort_InputInvalidException | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_SequenceTooShort_InputInvalidExceptionjalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_SequenceTooShort_InputInvalidException | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_EmptyColumnAndSubmitGaps_ColumnExtrudedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_EmptyColumnAndSubmitGaps_ColumnExtruded | 1PASS | ||
|
0.23333333
|
jalview.datamodel.AlignmentAnnotationTests.testAdjustForAlignmentjalview.datamodel.AlignmentAnnotationTests.testAdjustForAlignment | 1PASS | ||
|
0.23333333
|
jalview.gui.PairwiseAlignmentPanelTest.testConstructor_withSelectionGroupjalview.gui.PairwiseAlignmentPanelTest.testConstructor_withSelectionGroup | 1PASS | ||
|
0.23333333
|
jalview.util.MappingUtilsTest.testMapEditCommandjalview.util.MappingUtilsTest.testMapEditCommand | 1PASS | ||
|
0.23333333
|
jalview.io.JalviewExportPropertiesTests.testImportExportPeriodGapsjalview.io.JalviewExportPropertiesTests.testImportExportPeriodGaps | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistancesjalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances | 1PASS | ||
|
0.23333333
|
jalview.datamodel.AlignmentTest.testIsSecondaryStructurePresentjalview.datamodel.AlignmentTest.testIsSecondaryStructurePresent | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_EqualLengthNoGapsNoSubmitGapsjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_EqualLengthNoGapsNoSubmitGaps | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_transparencySingleFeaturejalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_transparencySingleFeature | 1PASS | ||
|
0.23333333
|
jalview.gui.PopupMenuTest.testConfigureReferenceAnnotationsMenu_alreadyAddedjalview.gui.PopupMenuTest.testConfigureReferenceAnnotationsMenu_alreadyAdded | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testReplacejalview.commands.EditCommandTest.testReplace | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.noDuplicatePdbMappingsMadejalview.project.Jalview2xmlTests.noDuplicatePdbMappingsMade | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testPAEsaveRestorejalview.project.Jalview2xmlTests.testPAEsaveRestore | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequence_ShortSequenceRemovedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequence_ShortSequenceRemoved | 1PASS | ||
|
0.23333333
|
jalview.datamodel.HiddenSequencesTest.testHideShowSequence_withHiddenRepSequencejalview.datamodel.HiddenSequencesTest.testHideShowSequence_withHiddenRepSequence | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentUtilsTests.testAddReferenceAnnotationsjalview.analysis.AlignmentUtilsTests.testAddReferenceAnnotations | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.io.StockholmFileTest.rfamFileIOjalview.io.StockholmFileTest.rfamFileIO | 1PASS | ||
|
0.23333333
|
jalview.gui.PopupMenuTest.testConfigureReferenceAnnotationsMenu_notOnAlignmentjalview.gui.PopupMenuTest.testConfigureReferenceAnnotationsMenu_notOnAlignment | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.gui.SeqCanvasTest.testClear_HighlightAndSelectionjalview.gui.SeqCanvasTest.testClear_HighlightAndSelection | 1PASS | ||
|
0.23333333
|
jalview.io.FeaturesFileTest.simpleGff3FileLoaderjalview.io.FeaturesFileTest.simpleGff3FileLoader | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testColourByAnnotScoresjalview.project.Jalview2xmlTests.testColourByAnnotScores | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.analysis.DnaTest.testTranslateCdna_simplejalview.analysis.DnaTest.testTranslateCdna_simple | 1PASS | ||
|
0.23333333
|
jalview.io.AnnotatedPDBFileInputTest.checkAnnotationWiringjalview.io.AnnotatedPDBFileInputTest.checkAnnotationWiring | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testUndo_deleteGapjalview.commands.EditCommandTest.testUndo_deleteGap | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testLeftRight_Justify_and_preserves_gapsjalview.commands.EditCommandTest.testLeftRight_Justify_and_preserves_gaps | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequenceAndSubmitGaps_ShortSeqRemovedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequenceAndSubmitGaps_ShortSeqRemoved | 1PASS | ||
|
0.23333333
|
jalview.io.JSONFileTest.testBioJSONRoundTripWithColourSchemeNonejalview.io.JSONFileTest.testBioJSONRoundTripWithColourSchemeNone | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistancesjalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances | 1PASS | ||
|
0.23333333
|
jalview.io.StockholmFileTest.pfamFileDataExtractionjalview.io.StockholmFileTest.pfamFileDataExtraction | 1PASS | ||
|
0.23333333
|
jalview.renderer.ContactGeometryTest.testCoverageofRangejalview.renderer.ContactGeometryTest.testCoverageofRange | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionTest.retrieveResult_verifySequencesAlignedjalview.ws2.actions.alignment.AlignmentActionTest.retrieveResult_verifySequencesAligned | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceTest.testDeleteChars_withGapsjalview.datamodel.SequenceTest.testDeleteChars_withGaps | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testCut_withFeaturesjalview.commands.EditCommandTest.testCut_withFeatures | 1PASS | ||
|
0.23333333
|
jalview.util.MappingUtilsTest.testMapSequenceGroup_regionjalview.util.MappingUtilsTest.testMapSequenceGroup_region | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.gui.AlignmentPanelTest.testPrintWrappedAlignmentjalview.gui.AlignmentPanelTest.testPrintWrappedAlignment | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_UnequalLengthNoGapsSubmitGapsjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_UnequalLengthNoGapsSubmitGaps | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_graduatedFeatureColourjalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_graduatedFeatureColour | 1PASS | ||
|
0.23333333
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentUtilsTests.testExpandContext_annotationjalview.analysis.AlignmentUtilsTests.testExpandContext_annotation | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.gui.PairwiseAlignmentPanelTest.testConstructor_noSelectionGroupjalview.gui.PairwiseAlignmentPanelTest.testConstructor_noSelectionGroup | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances_withGapjalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances_withGap | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionTest.submitSequences_submitGapsOn_verifySequencesSubmittedWithGapsjalview.ws2.actions.alignment.AlignmentActionTest.submitSequences_submitGapsOn_verifySequencesSubmittedWithGaps | 1PASS | ||
|
0.23333333
|
jalview.io.CrossRef2xmlTests.openCrossrefsForEnsemblTwicejalview.io.CrossRef2xmlTests.openCrossrefsForEnsemblTwice | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.gatherViewsHerejalview.project.Jalview2xmlTests.gatherViewsHere | 1PASS | ||
|
0.23333333
|
jalview.ext.jmol.JmolViewerTest.testAddStrToSingleSeqViewJMoljalview.ext.jmol.JmolViewerTest.testAddStrToSingleSeqViewJMol | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SearchResultsTest.testGetSequencesjalview.datamodel.SearchResultsTest.testGetSequences | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceTest.testFindFeaturesjalview.datamodel.SequenceTest.testFindFeatures | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequenceAndRequireAligned_ShortSeqRemovedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequenceAndRequireAligned_ShortSeqRemoved | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testPcaViewAssociationjalview.project.Jalview2xmlTests.testPcaViewAssociation | 1PASS | ||
|
0.23333333
|
jalview.analysis.FinderTest.testFind_ignoreHiddenColumnsjalview.analysis.FinderTest.testFind_ignoreHiddenColumns | 1PASS | ||
|
0.23333333
|
jalview.io.RNAMLfileTest.testRnamlSeqImportjalview.io.RNAMLfileTest.testRnamlSeqImport | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testSaveAndLoadAnnotationBasedTreejalview.project.Jalview2xmlTests.testSaveAndLoadAnnotationBasedTree | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.viewRefPdbAnnotationjalview.project.Jalview2xmlTests.viewRefPdbAnnotation | 1PASS | ||
|
0.23333333
|
jalview.ext.jmol.JmolParserTest.testFileParserjalview.ext.jmol.JmolParserTest.testFileParser | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureRendererTest.testFindFeaturesAtColumnjalview.renderer.seqfeatures.FeatureRendererTest.testFindFeaturesAtColumn | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_featureGroupNotDisplayedjalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_featureGroupNotDisplayed | 1PASS | ||
|
0.23333333
|
jalview.io.HMMFileTest.testParseModeljalview.io.HMMFileTest.testParseModel | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.gui.AlignmentPanelTest.testPrintWrappedAlignmentjalview.gui.AlignmentPanelTest.testPrintWrappedAlignment | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_SequenceGroup_RegionSubmittedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_SequenceGroup_RegionSubmitted | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_SequenceTooShort_InputInvalidExceptionjalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_SequenceTooShort_InputInvalidException | 1PASS | ||
|
0.23333333
|
jalview.ws.ebi.HmmerJSONProcessTest.parseJsonResultTestjalview.ws.ebi.HmmerJSONProcessTest.parseJsonResultTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequenceAndSubmitGaps_ShortSeqRemovedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequenceAndSubmitGaps_ShortSeqRemoved | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.submitSequences_submitGapsOff_verifySequencesSubmittedWithoutGapsjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.submitSequences_submitGapsOff_verifySequencesSubmittedWithoutGaps | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testCut_withFeatures_exhaustivejalview.commands.EditCommandTest.testCut_withFeatures_exhaustive | 1PASS | ||
|
0.23333333
|
jalview.ws.ebi.HmmerJSONProcessTest.parseHitTestjalview.ws.ebi.HmmerJSONProcessTest.parseHitTest | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceTest.testFindPositionsjalview.datamodel.SequenceTest.testFindPositions | 1PASS | ||
|
0.23333333
|
jalview.datamodel.AlignmentTest.testPropagateInsertionsOverlapjalview.datamodel.AlignmentTest.testPropagateInsertionsOverlap | 1PASS | ||
|
0.23333333
|
jalview.structures.models.AAStructureBindingModelTest.testImportPDBPreservesChainMappingsjalview.structures.models.AAStructureBindingModelTest.testImportPDBPreservesChainMappings | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testCopyViewSettingsjalview.project.Jalview2xmlTests.testCopyViewSettings | 1PASS | ||
|
0.23333333
|
jalview.gui.SeqPanelTest.testSetStatusReturnsNearestResiduePositionjalview.gui.SeqPanelTest.testSetStatusReturnsNearestResiduePosition | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.io.StockholmFileTest.pfamFileIOjalview.io.StockholmFileTest.pfamFileIO | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testUndo_insertGapjalview.commands.EditCommandTest.testUndo_insertGap | 1PASS | ||
|
0.23333333
|
jalview.datamodel.HiddenMarkovModelTest.testGetInsertEmissionProbabilitiesjalview.datamodel.HiddenMarkovModelTest.testGetInsertEmissionProbabilities | 1PASS | ||
|
0.23333333
|
jalview.util.MappingUtilsTest.testMapSequenceGroup_sequencesjalview.util.MappingUtilsTest.testMapSequenceGroup_sequences | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances_withSSUndefinedInBothSeqsjalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances_withSSUndefinedInBothSeqs | 1PASS | ||
|
0.23333333
|
jalview.gui.AlignmentPanelTest.testPrintWrappedAlignmentjalview.gui.AlignmentPanelTest.testPrintWrappedAlignment | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModel_HiddenColumnsjalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModel_HiddenColumns | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceTest.testDeleteCharsjalview.datamodel.SequenceTest.testDeleteChars | 1PASS | ||
|
0.23333333
|
jalview.structures.models.AAStructureBindingModelTest.testBuildColoursMapjalview.structures.models.AAStructureBindingModelTest.testBuildColoursMap | 1PASS | ||
|
0.23333333
|
jalview.gui.DesktopTests.testInternalCopyPastejalview.gui.DesktopTests.testInternalCopyPaste | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testSaveAndLoadAnnotationsPropertiesjalview.project.Jalview2xmlTests.testSaveAndLoadAnnotationsProperties | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testAutoShowOverviewForLegacyProjectsjalview.project.Jalview2xmlTests.testAutoShowOverviewForLegacyProjects | 1PASS | ||
|
0.23333333
|
jalview.gui.AnnotationColumnChooserTest.testResetjalview.gui.AnnotationColumnChooserTest.testReset | 1PASS | ||
|
0.23333333
|
jalview.analysis.AAFrequencyTest.testGetAnalogueCountjalview.analysis.AAFrequencyTest.testGetAnalogueCount | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_RegionIncludingGapsAndSubmitGaps_RegionSubmittedGapsExtrudedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_RegionIncludingGapsAndSubmitGaps_RegionSubmittedGapsExtruded | 1PASS | ||
|
0.23333333
|
jalview.io.JSONFileTest.testGrpParsed_colourNonejalview.io.JSONFileTest.testGrpParsed_colourNone | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModel_hiddenFirstColumnjalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModel_hiddenFirstColumn | 1PASS | ||
|
0.23333333
|
jalview.util.MappingUtilsTest.testMapSequenceGroup_columnsjalview.util.MappingUtilsTest.testMapSequenceGroup_columns | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.io.StockholmFileTest.fullWUSSsecondaryStructureForRNASequencejalview.io.StockholmFileTest.fullWUSSsecondaryStructureForRNASequence | 1PASS | ||
|
0.23333333
|
jalview.datamodel.AlignmentTest.testPropagateInsertionsjalview.datamodel.AlignmentTest.testPropagateInsertions | 1PASS | ||
|
0.23333333
|
jalview.datamodel.AlignmentViewTest.testGetVisibleAlignmentGapCharjalview.datamodel.AlignmentViewTest.testGetVisibleAlignmentGapChar | 1PASS | ||
|
0.23333333
|
jalview.gui.AlignFrameTest.testHideFeatureColumnsjalview.gui.AlignFrameTest.testHideFeatureColumns | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.submitSequences_verifySequenceNamesUniquifiedjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.submitSequences_verifySequenceNamesUniquified | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistancesjalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances | 1PASS | ||
|
0.23333333
|
jalview.gui.AnnotationLabelsTest2.testIdWidthNoChangesjalview.gui.AnnotationLabelsTest2.testIdWidthNoChanges | 1PASS | ||
|
0.23333333
|
jalview.gui.AlignmentPanelTest.testPrintWrappedAlignmentjalview.gui.AlignmentPanelTest.testPrintWrappedAlignment | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.allJobsStarted_taskStartedCalledjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.allJobsStarted_taskStartedCalled | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverPDBEntryjalview.project.Jalview2xmlTests.testStoreAndRecoverPDBEntry | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest | 1PASS | ||
|
0.23333333
|
jalview.controller.AlignViewControllerTest.testFindColumnsWithFeaturejalview.controller.AlignViewControllerTest.testFindColumnsWithFeature | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceGroupTest.testHasSecondaryStructureAnnotationjalview.datamodel.SequenceGroupTest.testHasSecondaryStructureAnnotation | 1PASS | ||
|
0.23333333
|
jalview.io.StockholmFileTest.secondaryStructureForRNASequencejalview.io.StockholmFileTest.secondaryStructureForRNASequence | 1PASS | ||
|
0.23333333
|
jalview.analysis.AAFrequencyTest.testExtractHMMProfilejalview.analysis.AAFrequencyTest.testExtractHMMProfile | 1PASS | ||
|
0.23333333
|
jalview.structures.models.AAStructureBindingModelTest.testFindSuperposableResiduesjalview.structures.models.AAStructureBindingModelTest.testFindSuperposableResidues | 1PASS | ||
|
0.23333333
|
jalview.datamodel.AlignmentAnnotationTests.testCopyConstructorjalview.datamodel.AlignmentAnnotationTests.testCopyConstructor | 1PASS | ||
|
0.23333333
|
jalview.gui.AnnotationLabelsTest2.testIdWidthNoChangesjalview.gui.AnnotationLabelsTest2.testIdWidthNoChanges | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.submitSequences_submitGapsOn_verifySequencesSubmittedWithGapsjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.submitSequences_submitGapsOn_verifySequencesSubmittedWithGaps | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverExpandedviewsjalview.project.Jalview2xmlTests.testStoreAndRecoverExpandedviews | 1PASS | ||
|
0.23333333
|
jalview.analysis.DnaTest.testTranslateCdna_hiddenColumnsjalview.analysis.DnaTest.testTranslateCdna_hiddenColumns | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_UnequalLengthNoGapsSubmitGapsRequireAlignedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_UnequalLengthNoGapsSubmitGapsRequireAligned | 1PASS | ||
|
0.23333333
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_UnequalLengthNoGapsNoSubmitGapsjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_UnequalLengthNoGapsNoSubmitGaps | 1PASS | ||
|
0.23333333
|
jalview.datamodel.PAEContactMatrixTest.testSeqAssociatedPAEMatrixjalview.datamodel.PAEContactMatrixTest.testSeqAssociatedPAEMatrix | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testReplaceFirstResiduesWithGapsjalview.commands.EditCommandTest.testReplaceFirstResiduesWithGaps | 1PASS | ||
|
0.23333333
|
jalview.gui.SeqPanelTest.testAmbiguousAminoAcidGetsStatusMessagejalview.gui.SeqPanelTest.testAmbiguousAminoAcidGetsStatusMessage | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentUtilsTests.testExpandContextjalview.analysis.AlignmentUtilsTests.testExpandContext | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SeqCigarTest.testSomethingjalview.datamodel.SeqCigarTest.testSomething | 1PASS | ||
|
0.23333333
|
jalview.ext.pymol.PymolManagerTest.testGetPostRequestjalview.ext.pymol.PymolManagerTest.testGetPostRequest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_NonStandardInColumnsAndFilterNonStandard_ColumnsExtrudedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_NonStandardInColumnsAndFilterNonStandard_ColumnsExtruded | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFindDistancesjalview.analysis.scoremodels.FeatureDistanceModelTest.testFindDistances | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_ContainsNonStandardAndFilterNonStandard_NonStandardToGapjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_ContainsNonStandardAndFilterNonStandard_NonStandardToGap | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_SequenceTooShort_InputInvalidExceptionjalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_SequenceTooShort_InputInvalidException | 1PASS | ||
|
0.23333333
|
jalview.commands.TrimRegionCommandTest.testTrimRight_oneColumnjalview.commands.TrimRegionCommandTest.testTrimRight_oneColumn | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testCollectResultsjalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testCollectResults | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testCollectResultsjalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testCollectResults | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistancesjalview.analysis.scoremodels.SecondaryStructureDistanceModelTest.testFindDistances | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testUndo_multipleInsertGapsjalview.commands.EditCommandTest.testUndo_multipleInsertGaps | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentUtilsTests.testAddMappedPositionsjalview.analysis.AlignmentUtilsTests.testAddMappedPositions | 1PASS | ||
|
0.23333333
|
jalview.datamodel.AlignmentAnnotationTests.testLiftOverjalview.datamodel.AlignmentAnnotationTests.testLiftOver | 1PASS | ||
|
0.23333333
|
jalview.util.MappingUtilsTest.testMapColumnSelection_dnaToProteinjalview.util.MappingUtilsTest.testMapColumnSelection_dnaToProtein | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequence_ShortSequenceRemovedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequence_ShortSequenceRemoved | 1PASS | ||
|
0.23333333
|
jalview.util.MappingUtilsTest.testMapSequenceGroup_sharedDatasetjalview.util.MappingUtilsTest.testMapSequenceGroup_sharedDataset | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModeljalview.analysis.scoremodels.FeatureDistanceModelTest.testFeatureScoreModel | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testUndo_multipleCommandsjalview.commands.EditCommandTest.testUndo_multipleCommands | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentUtilsTests.testAlignAsSameSequencesjalview.analysis.AlignmentUtilsTests.testAlignAsSameSequences | 1PASS | ||
|
0.23333333
|
jalview.controller.AlignViewControllerTest.testSelectColumnsWithHighlightjalview.controller.AlignViewControllerTest.testSelectColumnsWithHighlight | 1PASS | ||
|
0.23333333
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.23333333
|
jalview.renderer.ContactGeometryTest.testContactGeometryjalview.renderer.ContactGeometryTest.testContactGeometry | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverAnnotationRowElementColoursjalview.project.Jalview2xmlTests.testStoreAndRecoverAnnotationRowElementColours | 1PASS | ||
|
0.23333333
|
jalview.gui.AlignViewportTest.testGetSelectionAsNewSequences_withContactMatricesjalview.gui.AlignViewportTest.testGetSelectionAsNewSequences_withContactMatrices | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequence_ShortSequenceRemovedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequence_ShortSequenceRemoved | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceTest.testLocateVisibleStartofSequencejalview.datamodel.SequenceTest.testLocateVisibleStartofSequence | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_SequenceTooShort_InputInvalidExceptionjalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testPrepareJobs_SequenceTooShort_InputInvalidException | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentUtilsTests.testShowOrHideSequenceAnnotationsjalview.analysis.AlignmentUtilsTests.testShowOrHideSequenceAnnotations | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.23333333
|
jalview.gui.SeqPanelTest.testFindMousePosition_wrapped_scales_longSequencejalview.gui.SeqPanelTest.testFindMousePosition_wrapped_scales_longSequence | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureAtEndjalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureAtEnd | 1PASS | ||
|
0.23333333
|
jalview.datamodel.AlignmentViewTest.testGetVisibleContigsjalview.datamodel.AlignmentViewTest.testGetVisibleContigs | 1PASS | ||
|
0.23333333
|
jalview.structures.models.AAStructureBindingModelTest.testFindSuperposableResidues_hiddenColumnjalview.structures.models.AAStructureBindingModelTest.testFindSuperposableResidues_hiddenColumn | 1PASS | ||
|
0.23333333
|
jalview.io.RNAMLfileTest.testRnamlToStockholmIOjalview.io.RNAMLfileTest.testRnamlToStockholmIO | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceTest.testCopyPasteStyleDerivesequence_withcontactMatrixAnnjalview.datamodel.SequenceTest.testCopyPasteStyleDerivesequence_withcontactMatrixAnn | 1PASS | ||
|
0.23333333
|
jalview.analysis.FinderTest.testFind_withHiddenColumnsAndSelectionjalview.analysis.FinderTest.testFind_withHiddenColumnsAndSelection | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testAppendEdit_deleteGapjalview.commands.EditCommandTest.testAppendEdit_deleteGap | 1PASS | ||
|
0.23333333
|
jalview.analysis.DnaTest.testTranslateCdna_sequenceOrderIndependentjalview.analysis.DnaTest.testTranslateCdna_sequenceOrderIndependent | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_graduatedWithThresholdjalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_graduatedWithThreshold | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ext.jmol.JmolCommandsTest.testGetColourBySequenceCommands_hiddenColumnsjalview.ext.jmol.JmolCommandsTest.testGetColourBySequenceCommands_hiddenColumns | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SearchResultsTest.testMarkColumnsjalview.datamodel.SearchResultsTest.testMarkColumns | 1PASS | ||
|
0.23333333
|
jalview.datamodel.PAEContactMatrixTest.testMappableContactMatrixjalview.datamodel.PAEContactMatrixTest.testMappableContactMatrix | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.allJobsStarted_taskStatusChangedCalledWithReadyThenSubmittedjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.allJobsStarted_taskStatusChangedCalledWithReadyThenSubmitted | 1PASS | ||
|
0.23333333
|
jalview.schemes.HmmerGlobalBackgroundTest.testFindColourjalview.schemes.HmmerGlobalBackgroundTest.testFindColour | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentUtilsTests.testAddMappedPositions_withStopCodonjalview.analysis.AlignmentUtilsTests.testAddMappedPositions_withStopCodon | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.structure.StructureSelectionManagerTest.testSetMapping_seqFeaturesjalview.structure.StructureSelectionManagerTest.testSetMapping_seqFeatures | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_NonStandardInColumnsAndFilterNonStandard_ColumnsExtrudedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_NonStandardInColumnsAndFilterNonStandard_ColumnsExtruded | 1PASS | ||
|
0.23333333
|
jalview.analysis.AverageDistanceEngineTest.testUPGMAEnginejalview.analysis.AverageDistanceEngineTest.testUPGMAEngine | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequenceAndRequireAligned_ShortSeqRemovedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_TooShortSequenceAndRequireAligned_ShortSeqRemoved | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_ContainsNonStandardAndNoFilterNonStandard_NonStandardRemainjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_ContainsNonStandardAndNoFilterNonStandard_NonStandardRemain | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testRNAStructureRecoveryjalview.project.Jalview2xmlTests.testRNAStructureRecovery | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceTest.testDeriveSequence_existingDatasetjalview.datamodel.SequenceTest.testDeriveSequence_existingDataset | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_singleFeatureAtPositionjalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_singleFeatureAtPosition | 1PASS | ||
|
0.23333333
|
jalview.analysis.FinderTest.testFind_residueNumberjalview.analysis.FinderTest.testFind_residueNumber | 1PASS | ||
|
0.23333333
|
jalview.gui.AssociatePDBFileTest.testAssociatePDBFilejalview.gui.AssociatePDBFileTest.testAssociatePDBFile | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentUtilsTests.testAlignAsSameSequencesMultipleSubSeqjalview.analysis.AlignmentUtilsTests.testAlignAsSameSequencesMultipleSubSeq | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_nestedFeaturesjalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_nestedFeatures | 1PASS | ||
|
0.23333333
|
jalview.analysis.FinderTest.testFindAll_inSelectionjalview.analysis.FinderTest.testFindAll_inSelection | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceTest.testGetSubsequencejalview.datamodel.SequenceTest.testGetSubsequence | 1PASS | ||
|
0.23333333
|
jalview.schemes.HmmerLocalBackgroundTest.testFindColourjalview.schemes.HmmerLocalBackgroundTest.testFindColour | 1PASS | ||
|
0.23333333
|
jalview.commands.TrimRegionCommandTest.testTrimRight_withUndoAndRedojalview.commands.TrimRegionCommandTest.testTrimRight_withUndoAndRedo | 1PASS | ||
|
0.23333333
|
jalview.io.HMMFileTest.testParsejalview.io.HMMFileTest.testParse | 1PASS | ||
|
0.23333333
|
jalview.io.HMMFileTest.testParse_minimalFilejalview.io.HMMFileTest.testParse_minimalFile | 1PASS | ||
|
0.23333333
|
jalview.util.MappingUtilsTest.testMapColumnSelection_hiddenColumnsjalview.util.MappingUtilsTest.testMapColumnSelection_hiddenColumns | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_network_structureOpeningArgsTestjalview.bin.CommandsTest2.do_network_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.analysis.FinderTest.testFind_featuresOnlyjalview.analysis.FinderTest.testFind_featuresOnly | 1PASS | ||
|
0.23333333
|
mc_view.PDBChainTest.testMakeResidueList_withTempFactormc_view.PDBChainTest.testMakeResidueList_withTempFactor | 1PASS | ||
|
0.23333333
|
jalview.gui.AnnotationLabelsTest2.testIdWidthChangesjalview.gui.AnnotationLabelsTest2.testIdWidthChanges | 1PASS | ||
|
0.23333333
|
jalview.analysis.FinderTest.testFind_withHiddenColumnsjalview.analysis.FinderTest.testFind_withHiddenColumns | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.datamodel.AlignmentTest.testPadGapsjalview.datamodel.AlignmentTest.testPadGaps | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_network_structureOpeningArgsTestjalview.bin.CommandsTest2.do_network_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.datamodel.AlignmentTest.testAlignAs_dnaAsDnajalview.datamodel.AlignmentTest.testAlignAs_dnaAsDna | 1PASS | ||
|
0.23333333
|
mc_view.PDBfileTest.testParse_withAnnotations_noSSmc_view.PDBfileTest.testParse_withAnnotations_noSS | 1PASS | ||
|
0.23333333
|
jalview.commands.TrimRegionCommandTest.testTrimLeft_oneColumnjalview.commands.TrimRegionCommandTest.testTrimLeft_oneColumn | 1PASS | ||
|
0.23333333
|
jalview.analysis.DnaTest.testTranslateCdna_withUntranslatableCodonsjalview.analysis.DnaTest.testTranslateCdna_withUntranslatableCodons | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionTest.submitSequences_submitGapsOff_verifySequencesSubmittedWithoutGapsjalview.ws2.actions.alignment.AlignmentActionTest.submitSequences_submitGapsOff_verifySequencesSubmittedWithoutGaps | 1PASS | ||
|
0.23333333
|
jalview.gui.AnnotationLabelsTest2.testIdWidthChangesjalview.gui.AnnotationLabelsTest2.testIdWidthChanges | 1PASS | ||
|
0.23333333
|
jalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_singleFeatureNotAtPositionjalview.renderer.seqfeatures.FeatureColourFinderTest.testFindFeatureColour_singleFeatureNotAtPosition | 1PASS | ||
|
0.23333333
|
jalview.analysis.scoremodels.FeatureDistanceModelTest.testFindFeatureAt_PointFeaturejalview.analysis.scoremodels.FeatureDistanceModelTest.testFindFeatureAt_PointFeature | 1PASS | ||
|
0.23333333
|
jalview.datamodel.CigarArrayTest.testConstructorjalview.datamodel.CigarArrayTest.testConstructor | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.analysis.DnaTest.testTranslateCdna_withUntranslatableCodonsAndHiddenColumnsjalview.analysis.DnaTest.testTranslateCdna_withUntranslatableCodonsAndHiddenColumns | 1PASS | ||
|
0.23333333
|
jalview.io.FeaturesFileTest.simpleGff3RelaxedIdMatchingjalview.io.FeaturesFileTest.simpleGff3RelaxedIdMatching | 1PASS | ||
|
0.23333333
|
jalview.datamodel.HiddenMarkovModelTest.testGetMatchEmissionProbabilitiesjalview.datamodel.HiddenMarkovModelTest.testGetMatchEmissionProbabilities | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_taskStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.gui.PopupMenuTest.testConfigureReferenceAnnotationsMenujalview.gui.PopupMenuTest.testConfigureReferenceAnnotationsMenu | 1PASS | ||
|
0.23333333
|
jalview.datamodel.HiddenMarkovModelTest.testGetStateTransitionProbabilitiesjalview.datamodel.HiddenMarkovModelTest.testGetStateTransitionProbabilities | 1PASS | ||
|
0.23333333
|
jalview.analysis.AAFrequencyTest.testCompleteInformationjalview.analysis.AAFrequencyTest.testCompleteInformation | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SequenceTest.testDeleteChars_withDbRefsAndFeaturesjalview.datamodel.SequenceTest.testDeleteChars_withDbRefsAndFeatures | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testCollectResultjalview.ws2.actions.secstructpred.SecStructPredPDBSearchTaskTest.testCollectResult | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionTest.submitSequences_verifySequenceNamesUniquifiedjalview.ws2.actions.alignment.AlignmentActionTest.submitSequences_verifySequenceNamesUniquified | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatusjalview.ws2.actions.alignment.AlignmentActionListenerNotifiedTest.jobStatusChanged_subJobStatusChangedCalledWithJobStatus | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentUtilsTests.testAlignAs_alternateTranscriptsUngappedjalview.analysis.AlignmentUtilsTests.testAlignAs_alternateTranscriptsUngapped | 1PASS | ||
|
0.23333333
|
jalview.analysis.AlignmentUtilsTests.testAddReferenceContactMapjalview.analysis.AlignmentUtilsTests.testAddReferenceContactMap | 1PASS | ||
|
0.23333333
|
jalview.util.MappingUtilsTest.testMapColumnSelection_proteinToDnajalview.util.MappingUtilsTest.testMapColumnSelection_proteinToDna | 1PASS | ||
|
0.23333333
|
jalview.project.Jalview2xmlTests.testTCoffeeScoresjalview.project.Jalview2xmlTests.testTCoffeeScores | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testUndo_cutjalview.commands.EditCommandTest.testUndo_cut | 1PASS | ||
|
0.23333333
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest | 1PASS | ||
|
0.23333333
|
jalview.datamodel.HiddenMarkovModelTest.testGetInformationContentjalview.datamodel.HiddenMarkovModelTest.testGetInformationContent | 1PASS | ||
|
0.23333333
|
jalview.analysis.AAFrequencyTest.testExtractSSProfileForSequenceGroupjalview.analysis.AAFrequencyTest.testExtractSSProfileForSequenceGroup | 1PASS | ||
|
0.23333333
|
jalview.datamodel.SeqCigarTest.testFindPositionjalview.datamodel.SeqCigarTest.testFindPosition | 1PASS | ||
|
0.23333333
|
jalview.io.AnnotatedPDBFileInputTest.checkNoDuplicatesjalview.io.AnnotatedPDBFileInputTest.checkNoDuplicates | 1PASS | ||
|
0.23333333
|
jalview.commands.EditCommandTest.testCut_withFeatures5primejalview.commands.EditCommandTest.testCut_withFeatures5prime | 1PASS | ||
|
0.23333333
|
mc_view.PDBfileTest.testParse_withJmol_noAnnotationsmc_view.PDBfileTest.testParse_withJmol_noAnnotations | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequencesjalview.ws2.actions.secstructpred.SecStructPredMsaTaskTest.testPrepareJobs_checkInputSequences | 1PASS | ||
|
0.23333333
|
jalview.bin.CommandsTest2.do_structureOpeningArgsTestjalview.bin.CommandsTest2.do_structureOpeningArgsTest | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_EqualLengthNoGapsSubmitGapsjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_EqualLengthNoGapsSubmitGaps | 1PASS | ||
|
0.23333333
|
jalview.ws2.actions.annotation.AnnotationJobTest.testCreate_EmptyColumnAndRequireAligned_ColumnExtrudedjalview.ws2.actions.annotation.AnnotationJobTest.testCreate_EmptyColumnAndRequireAligned_ColumnExtruded | 1PASS |
Source view
| 1 | /* | ||||||
| 2 | * Jalview - A Sequence Alignment Editor and Viewer ($$Version-Rel$$) | ||||||
| 3 | * Copyright (C) $$Year-Rel$$ The Jalview Authors | ||||||
| 4 | * | ||||||
| 5 | * This file is part of Jalview. | ||||||
| 6 | * | ||||||
| 7 | * Jalview is free software: you can redistribute it and/or | ||||||
| 8 | * modify it under the terms of the GNU General Public License | ||||||
| 9 | * as published by the Free Software Foundation, either version 3 | ||||||
| 10 | * of the License, or (at your option) any later version. | ||||||
| 11 | * | ||||||
| 12 | * Jalview is distributed in the hope that it will be useful, but | ||||||
| 13 | * WITHOUT ANY WARRANTY; without even the implied warranty | ||||||
| 14 | * of MERCHANTABILITY or FITNESS FOR A PARTICULAR | ||||||
| 15 | * PURPOSE. See the GNU General Public License for more details. | ||||||
| 16 | * | ||||||
| 17 | * You should have received a copy of the GNU General Public License | ||||||
| 18 | * along with Jalview. If not, see <http://www.gnu.org/licenses/>. | ||||||
| 19 | * The Jalview Authors are detailed in the 'AUTHORS' file. | ||||||
| 20 | */ | ||||||
| 21 | package jalview.datamodel; | ||||||
| 22 | |||||||
| 23 | /** | ||||||
| 24 | * An immutable object representing one or more residue and corresponding | ||||||
| 25 | * alignment column positions for a sequence | ||||||
| 26 | */ | ||||||
|
|||||||
| 27 | public class SequenceCursor | ||||||
| 28 | { | ||||||
| 29 | /** | ||||||
| 30 | * the aligned sequence this cursor applies to | ||||||
| 31 | */ | ||||||
| 32 | public final SequenceI sequence; | ||||||
| 33 | |||||||
| 34 | /** | ||||||
| 35 | * residue position in sequence (start...), 0 if undefined | ||||||
| 36 | */ | ||||||
| 37 | public final int residuePosition; | ||||||
| 38 | |||||||
| 39 | /** | ||||||
| 40 | * column position (1...) corresponding to residuePosition, or 0 if undefined | ||||||
| 41 | */ | ||||||
| 42 | public final int columnPosition; | ||||||
| 43 | |||||||
| 44 | /** | ||||||
| 45 | * column position (1...) of first residue in the sequence, or 0 if undefined | ||||||
| 46 | */ | ||||||
| 47 | public final int firstColumnPosition; | ||||||
| 48 | |||||||
| 49 | /** | ||||||
| 50 | * column position (1...) of last residue in the sequence, or 0 if undefined | ||||||
| 51 | */ | ||||||
| 52 | public final int lastColumnPosition; | ||||||
| 53 | |||||||
| 54 | /** | ||||||
| 55 | * a token which may be used to check whether this cursor is still valid for | ||||||
| 56 | * its sequence (allowing it to be ignored if the sequence has changed) | ||||||
| 57 | */ | ||||||
| 58 | public final int token; | ||||||
| 59 | |||||||
| 60 | /** | ||||||
| 61 | * Constructor | ||||||
| 62 | * | ||||||
| 63 | * @param seq | ||||||
| 64 | * sequence this cursor applies to | ||||||
| 65 | * @param resPos | ||||||
| 66 | * residue position in sequence (start..) | ||||||
| 67 | * @param column | ||||||
| 68 | * column position in alignment (1..) | ||||||
| 69 | * @param tok | ||||||
| 70 | * a token that may be validated by the sequence to check the cursor | ||||||
| 71 | * is not stale | ||||||
| 72 | */ | ||||||
|
|||||||
| 73 | 25 |  public SequenceCursor(SequenceI seq, int resPos, int column, int tok)... public SequenceCursor(SequenceI seq, int resPos, int column, int tok)... |
|||||
| 74 | { | ||||||
| 75 | 25 | this(seq, resPos, column, 0, 0, tok); | |||||
| 76 | } | ||||||
| 77 | |||||||
| 78 | /** | ||||||
| 79 | * Constructor | ||||||
| 80 | * | ||||||
| 81 | * @param seq | ||||||
| 82 | * sequence this cursor applies to | ||||||
| 83 | * @param resPos | ||||||
| 84 | * residue position in sequence (start..) | ||||||
| 85 | * @param column | ||||||
| 86 | * column position in alignment (1..) | ||||||
| 87 | * @param firstResCol | ||||||
| 88 | * column position of the first residue in the sequence (1..), or 0 | ||||||
| 89 | * if not known | ||||||
| 90 | * @param lastResCol | ||||||
| 91 | * column position of the last residue in the sequence (1..), or 0 if | ||||||
| 92 | * not known | ||||||
| 93 | * @param tok | ||||||
| 94 | * a token that may be validated by the sequence to check the cursor | ||||||
| 95 | * is not stale | ||||||
| 96 | */ | ||||||
|
|||||||
| 97 | 1081678 |  public SequenceCursor(SequenceI seq, int resPos, int column,... public SequenceCursor(SequenceI seq, int resPos, int column,... |
|||||
| 98 | int firstResCol, int lastResCol, int tok) | ||||||
| 99 | { | ||||||
| 100 | 1081679 | sequence = seq; | |||||
| 101 | 1081686 | residuePosition = resPos; | |||||
| 102 | 1081701 | columnPosition = column; | |||||
| 103 | 1081702 | firstColumnPosition = firstResCol; | |||||
| 104 | 1081734 | lastColumnPosition = lastResCol; | |||||
| 105 | 1081740 | token = tok; | |||||
| 106 | } | ||||||
| 107 | |||||||
|
|||||||
| 108 | 0 |  @Override... @Override... |
|||||
| 109 | public int hashCode() | ||||||
| 110 | { | ||||||
| 111 | 0 | int hash = 31 * residuePosition; | |||||
| 112 | 0 | hash = 31 * hash + columnPosition; | |||||
| 113 | 0 | hash = 31 * hash + token; | |||||
| 114 | 0 | if (sequence != null) | |||||
| 115 | { | ||||||
| 116 | 0 | hash += sequence.hashCode(); | |||||
| 117 | } | ||||||
| 118 | 0 | return hash; | |||||
| 119 | } | ||||||
| 120 | |||||||
| 121 | /** | ||||||
| 122 | * Two cursors are equal if they refer to the same sequence object and have | ||||||
| 123 | * the same residue position, column position and token value | ||||||
| 124 | */ | ||||||
|
|||||||
| 125 | 13 |  @Override... @Override... |
|||||
| 126 | public boolean equals(Object obj) | ||||||
| 127 | { | ||||||
| 128 | 13 | if (!(obj instanceof SequenceCursor)) | |||||
| 129 | { | ||||||
| 130 | 0 | return false; | |||||
| 131 | } | ||||||
| 132 | 13 | SequenceCursor sc = (SequenceCursor) obj; | |||||
| 133 | 13 | return sequence == sc.sequence && residuePosition == sc.residuePosition | |||||
| 134 | && columnPosition == sc.columnPosition && token == sc.token; | ||||||
| 135 | } | ||||||
| 136 | |||||||
|
|||||||
| 137 | 31 |  @Override... @Override... |
|||||
| 138 | public String toString() | ||||||
| 139 | { | ||||||
| 140 | 31 | String name = sequence == null ? "" : sequence.getName(); | |||||
| 141 | 31 | return String.format("%s:Pos%d:Col%d:startCol%d:endCol%d:tok%d", name, | |||||
| 142 | residuePosition, columnPosition, firstColumnPosition, | ||||||
| 143 | lastColumnPosition, token); | ||||||
| 144 | } | ||||||
| 145 | } |