|
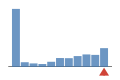
0.7297297
|
jalview.ext.jmol.JmolParserTest.testAlignmentLoaderjalview.ext.jmol.JmolParserTest.testAlignmentLoader
|
1PASS
|
|
|
0.6216216
|
jalview.bin.CommandsTest.structureImageOutputTestjalview.bin.CommandsTest.structureImageOutputTest
|
1PASS
|
|
|
0.6216216
|
jalview.bin.CommandsTest.structureImageOutputTestjalview.bin.CommandsTest.structureImageOutputTest
|
1PASS
|
|
|
0.5405405
|
mc_view.PDBfileTest.testParse_withJmol_noAnnotationsmc_view.PDBfileTest.testParse_withJmol_noAnnotations
|
1PASS
|
|
|
0.5405405
|
mc_view.PDBfileTest.testParse_withJmolAddAlignmentAnnotationsmc_view.PDBfileTest.testParse_withJmolAddAlignmentAnnotations
|
1PASS
|
|
|
0.45945945
|
jalview.io.AnnotatedPDBFileInputTest.testJalviewProjectRelocationAnnotationjalview.io.AnnotatedPDBFileInputTest.testJalviewProjectRelocationAnnotation
|
1PASS
|
|
|
0.45945945
|
jalview.ext.jmol.JmolViewerTest.testAddStrToSingleSeqViewJMoljalview.ext.jmol.JmolViewerTest.testAddStrToSingleSeqViewJMol
|
1PASS
|
|
|
0.45945945
|
jalview.ext.jmol.JmolViewerTest.testSingleSeqViewJMoljalview.ext.jmol.JmolViewerTest.testSingleSeqViewJMol
|
1PASS
|
|
|
0.45945945
|
jalview.structures.models.AAStructureBindingModelTest.testImportPDBPreservesChainMappingsjalview.structures.models.AAStructureBindingModelTest.testImportPDBPreservesChainMappings
|
1PASS
|
|
|
0.45945945
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverPDBEntryjalview.project.Jalview2xmlTests.testStoreAndRecoverPDBEntry
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest.structureImageOutputTestjalview.bin.CommandsTest.structureImageOutputTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest.structureImageOutputTestjalview.bin.CommandsTest.structureImageOutputTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest.structureImageAnnotationsOutputTestjalview.bin.CommandsTest.structureImageAnnotationsOutputTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest
|
1PASS
|
|
|
0.43243244
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest
|
1PASS
|
|
|
0.3783784
|
jalview.project.Jalview2xmlTests.noDuplicatePdbMappingsMadejalview.project.Jalview2xmlTests.noDuplicatePdbMappingsMade
|
1PASS
|
|
|
0.3783784
|
jalview.project.Jalview2xmlTests.viewRefPdbAnnotationjalview.project.Jalview2xmlTests.viewRefPdbAnnotation
|
1PASS
|
|
|
0.27027026
|
jalview.project.Jalview2xmlTests.testCopyViewSettingsjalview.project.Jalview2xmlTests.testCopyViewSettings
|
1PASS
|
|
|
0.27027026
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverExpandedviewsjalview.project.Jalview2xmlTests.testStoreAndRecoverExpandedviews
|
1PASS
|
|
|
0.27027026
|
jalview.ext.jmol.JmolParserTest.testParse_missingResiduesjalview.ext.jmol.JmolParserTest.testParse_missingResidues
|
1PASS
|
|
|
0.27027026
|
jalview.project.Jalview2xmlTests.testStoreAndRecoverReferenceSeqSettingsjalview.project.Jalview2xmlTests.testStoreAndRecoverReferenceSeqSettings
|
1PASS
|
|
|
0.27027026
|
jalview.ext.jmol.JmolParserTest.testFileParserjalview.ext.jmol.JmolParserTest.testFileParser
|
1PASS
|
|
|
0.27027026
|
jalview.project.Jalview2xmlTests.gatherViewsHerejalview.project.Jalview2xmlTests.gatherViewsHere
|
1PASS
|
|
|
0.27027026
|
jalview.structure.StructureSelectionManagerTest.testSetMapping_seqFeaturesjalview.structure.StructureSelectionManagerTest.testSetMapping_seqFeatures
|
1PASS
|
|
|
0.27027026
|
jalview.ext.jmol.JmolParserTest.testLocalPDBIdjalview.ext.jmol.JmolParserTest.testLocalPDBId
|
1PASS
|
|
|
0.27027026
|
jalview.ext.jmol.JmolParserTest.testParse_alternativeResiduesjalview.ext.jmol.JmolParserTest.testParse_alternativeResidues
|
1PASS
|
|
|
0.21621622
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest
|
1PASS
|
|
|
0.21621622
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest
|
1PASS
|
|
|
0.21621622
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest
|
1PASS
|
|
|
0.21621622
|
jalview.bin.CommandsTest2.structureOpeningArgsTestjalview.bin.CommandsTest2.structureOpeningArgsTest
|
1PASS
|
|
|
0.21621622
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest
|
1PASS
|
|
|
0.21621622
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest
|
1PASS
|
|
|
0.21621622
|
jalview.gui.StructureChooserTest.openStructureFileForSequenceTestjalview.gui.StructureChooserTest.openStructureFileForSequenceTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTestjalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTest
|
1PASS
|
|
|
0.10810811
|
jalview.gui.AnnotationLabelsTest2.testIdWidthNoChangesjalview.gui.AnnotationLabelsTest2.testIdWidthNoChanges
|
1PASS
|
|
|
0.10810811
|
jalview.io.BackupFilesTest.backupsEnabledRollMaxTestjalview.io.BackupFilesTest.backupsEnabledRollMaxTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTestjalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTestjalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTest
|
1PASS
|
|
|
0.10810811
|
jalview.util.imagemaker.BitmapImageSizeTest.testCacheSettingsRecoveryjalview.util.imagemaker.BitmapImageSizeTest.testCacheSettingsRecovery
|
1PASS
|
|
|
0.10810811
|
jalview.renderer.seqfeatures.FeatureRendererTest.testFindComplementFeaturesAtResiduejalview.renderer.seqfeatures.FeatureRendererTest.testFindComplementFeaturesAtResidue
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.io.BackupFilesTest.backupsEnabledSingleFileBackupTestjalview.io.BackupFilesTest.backupsEnabledSingleFileBackupTest
|
1PASS
|
|
|
0.10810811
|
jalview.ws.rest.RestClientTest.testGetRestClientjalview.ws.rest.RestClientTest.testGetRestClient
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.gui.DesktopTests.testInternalCopyPastejalview.gui.DesktopTests.testInternalCopyPaste
|
1PASS
|
|
|
0.10810811
|
jalview.gui.AnnotationLabelsTest2.testIdWidthChangesjalview.gui.AnnotationLabelsTest2.testIdWidthChanges
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CacheTest.testVersionCheckerjalview.bin.CacheTest.testVersionChecker
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTestjalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.gui.AnnotationLabelsTest2.testIdWidthChangesjalview.gui.AnnotationLabelsTest2.testIdWidthChanges
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.io.BackupFilesTest.noBackupsEnabledTestjalview.io.BackupFilesTest.noBackupsEnabledTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTestjalview.bin.CommandsTest.argFilesGlobAndSubstitutionsTest
|
1PASS
|
|
|
0.10810811
|
jalview.gui.AnnotationLabelsTest2.testIdWidthNoChangesjalview.gui.AnnotationLabelsTest2.testIdWidthNoChanges
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.io.BackupFilesTest.backupsEnabledNoRollMaxTestjalview.io.BackupFilesTest.backupsEnabledNoRollMaxTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.allLinkedIdsTestjalview.bin.CommandsTest.allLinkedIdsTest
|
1PASS
|
|
|
0.10810811
|
jalview.util.ImageMakerTest.testParseScaleWidthHeightStringsjalview.util.ImageMakerTest.testParseScaleWidthHeightStrings
|
1PASS
|
|
|
0.10810811
|
jalview.bin.CommandsTest.commandsOpenTestjalview.bin.CommandsTest.commandsOpenTest
|
1PASS
|
|
|
0.10810811
|
jalview.io.BackupFilesTest.backupsEnabledReverseRollMaxTestjalview.io.BackupFilesTest.backupsEnabledReverseRollMaxTest
|
1PASS
|
|
|
0.054054055
|
mc_view.PDBChainTest.testMakeExactMappingmc_view.PDBChainTest.testMakeExactMapping
|
1PASS
|
|
|
0.054054055
|
mc_view.PDBfileTest.testParse_withAnnotations_noSSmc_view.PDBfileTest.testParse_withAnnotations_noSS
|
1PASS
|
|
|
0.054054055
|
jalview.gui.AssociatePDBFileTest.testAssociatePDBFilejalview.gui.AssociatePDBFileTest.testAssociatePDBFile
|
1PASS
|
|
|
0.054054055
|
mc_view.PDBChainTest.testMakeResidueList_noAnnotationmc_view.PDBChainTest.testMakeResidueList_noAnnotation
|
1PASS
|
|
|
0.054054055
|
mc_view.PDBChainTest.testMakeCaBondList_nucleotidemc_view.PDBChainTest.testMakeCaBondList_nucleotide
|
1PASS
|
|
|
0.054054055
|
mc_view.PDBChainTest.testMakeResidueList_withTempFactormc_view.PDBChainTest.testMakeResidueList_withTempFactor
|
1PASS
|
|
|
0.054054055
|
mc_view.PDBfileTest.testParsemc_view.PDBfileTest.testParse
|
1PASS
|
|
|
0.054054055
|
jalview.gui.StructureChooserTest.fetchStructuresInfoMockedTestjalview.gui.StructureChooserTest.fetchStructuresInfoMockedTest
|
1PASS
|
|
|
0.054054055
|
jalview.gui.StructureChooserTest.populateFilterComboBoxTestjalview.gui.StructureChooserTest.populateFilterComboBoxTest
|
1PASS
|
|
|
0.054054055
|
jalview.gui.StructureChooserTest.displayTDBQueryTestjalview.gui.StructureChooserTest.displayTDBQueryTest
|
1PASS
|
|
|
0.054054055
|
mc_view.PDBChainTest.testMakeCaBondListmc_view.PDBChainTest.testMakeCaBondList
|
1PASS
|
|

 @Override
@Override public static void addSettings(boolean addAlignmentAnnotations,
public static void addSettings(boolean addAlignmentAnnotations, public static boolean isVisibleChainAnnotation()
public static boolean isVisibleChainAnnotation() public static void setTemperatureFactorType(TFType t)
public static void setTemperatureFactorType(TFType t) public static void setVisibleChainAnnotation(
public static void setVisibleChainAnnotation( public static boolean isProcessSecondaryStructure()
public static boolean isProcessSecondaryStructure() public static void setProcessSecondaryStructure(
public static void setProcessSecondaryStructure( public static boolean isExternalSecondaryStructure()
public static boolean isExternalSecondaryStructure() public static void setExternalSecondaryStructure(
public static void setExternalSecondaryStructure( public static boolean isShowSeqFeatures()
public static boolean isShowSeqFeatures() public static void setShowSeqFeatures(boolean showSeqFeatures)
public static void setShowSeqFeatures(boolean showSeqFeatures) public static PDBEntry.Type getDefaultStructureFileFormat()
public static PDBEntry.Type getDefaultStructureFileFormat() public static void setDefaultStructureFileFormat(
public static void setDefaultStructureFileFormat( public static String getDefaultPDBFileParser()
public static String getDefaultPDBFileParser() public static void setDefaultPDBFileParser(
public static void setDefaultPDBFileParser( public static void setDefaultPDBFileParser(String defaultPDBFileParser)
public static void setDefaultPDBFileParser(String defaultPDBFileParser) public static TFType getTemperatureFactorType()
public static TFType getTemperatureFactorType()